The Phyletica Lab has received an award from the National Science Foundation to develop a new statistical framework for inferring divergences shared across evolutionary lineages by generalizing Bayesian phylogenetics. Current phylogenetic models make an assumption: processes of diversification are strictly bifurcating and affect each evolutionary lineage independently. For example, think of a birth-death model in which each ‘birth’ event causes a single branch to split into two branches. When such an event occurs, it cannot affect any other contemporary branches across the phylogeny.
There are many, likely common, diversification processes that violate this assumption. Here are a few examples:
- Biogeography
- Geological or climatic changes to the landscape that affect entire communities of species
- Gene family evolution
- Chromosomal duplications that duplicate multiple members of a gene family
- Epidemiology
- Disease spread via co-infected individuals or simultaneous transmission at social gatherings
- Symbiont evolution
- Co-colonization of a new host
- Speciation of the host
All of these interesting scenarios can potentially generate divergences across phylogenies that are not independent, but rather temporally clustered.
Our goal is to accommodate such processes by generalizing the space of phylogenies considered during inference. Rather than restrict ourselves to considering the most parameter-rich class of phylogenetic models, in which all internal nodes are bifurcating and independent of one another (conditional on the topology), let’s consider all possible parameterizations of the tree (the figure below tries to illustrate this space of trees).
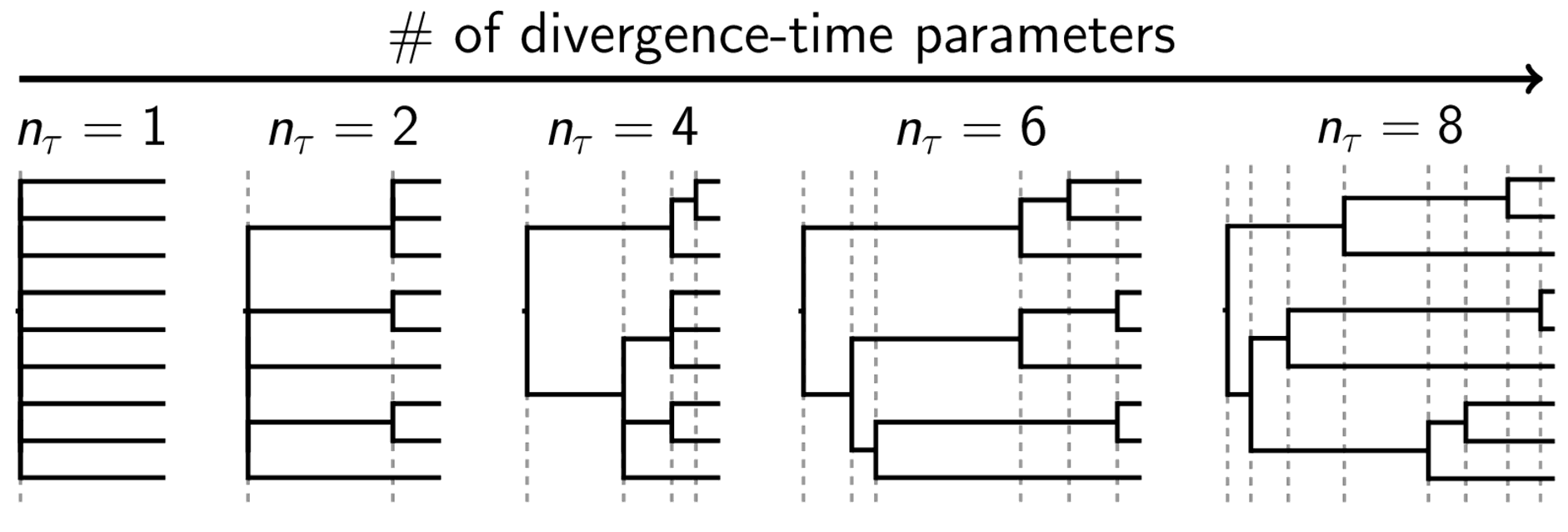
A sample of general tree space.
By allowing tree models with fewer divergence-time parameters onto the ‘playing field’ of phylogeny estimation, we will have a statistical framework for learning (and testing hypotheses) about processes of diversification that predict clusters of divergences across the tree of life. We will use this new method to infer the phylogeny of three genera of geckos that have all been evolving on the dynamic landscape of Southeast Asia for millions of years. The new method will allow us to test whether bouts of fragmentation of this landscape caused multiple gecko lineages to co-diverge. This work would not be possible without the help of great collaborators Perry Wood, Jr., Lee Grismer, and Cameron Siler.
